Some gut microbes that have coexisted with humans for a long time diversifies, a process known as codiversification. While gut microbes affect our immune system and health, it is unclear which species have coevolved with humans and to what extent. Coevolution is the evolution of organisms sharing the same environment in ways that affect each other, and coevolved microbes are important, because they provide clues not only for discovering more about human evolution, but also for exploring the involvement of gut microbes in health and disease. Exploring codiversified microbial species is the first step in seeking coevolved microbial species.
Special Feature 1 – Forefront Research into Gut Bacteria Exploring codiversified microbial species could lead to disease prevention or treatment
composition by Rie Iizuka
In the process of evolution, organisms that share an environment may affect each other. The term used to describe this in evolutionary biology is “coevolution.” Examples of such symbiotic systems are often seen among insects and invertebrates. Aphids suck the sap from plants, but they are unable to obtain the nutrients they require for survival from the sap in its original form. Accordingly, symbiotic bacteria in the aphid’s body break down the nectar and supply it with amino acids and vitamins. This coevolution has a very long history, to the extent that aphids and these microbes cannot live independently of each other. Consequently, aphid mothers pass on both their own genes and these symbiotic bacteria to their offspring. This process is similar to the way in which bacteria assimilated into eukaryotes back in ancient times exist in humans today in the form of mitochondria.
Coevolved microbes are important for learning about human evolution
Did humans and gut microbes coevolved? The gut microbiota consists of hundreds of microbial species that live in the human gut and affect humans’ immune systems and health. However, we do not have a good understanding of which species coevolved and to what extent. Coevolved microbes are likely to be important both for learning about human evolution and developing new treatments. Microbes have been passed down through countless generations from mother to child. The starting point for finding coevolved microbial species is searching for codiversified microbial species, where both microbes and humans have diversified sharing the same environment and history. Although coevolution and codiversification are similar concepts, coevolution differs from codiversification in that natural selection is an essential element of the former, so different terms are used. By decoding their respective genomes and comparing them from an evolutionary biology perspective, we can shed light on the codiversification of gut microbes and humans.
Research into lactose brought to light the relationship between gut microbes and human genes.The effects of human genes provide one of the mechanisms via which codiversified microbes are maintained in a stable state down through the generations. For example, an individual with lactose intolerance lacks the digestive enzyme lactase, which is secreted from the small intestine, with the result that lactose cannot be broken down. Consequently, that individual will suffer diarrhea and other symptoms if they consume lactose. Researchers in numerous countries have confirmed that the abundance of Bifidobacterium in the gut can be predcited by looking at the genotype of the lactase (LCT) gene, which is responsible for producing lactase.
If an individual has the lactose intolerance genotype, that means lactase is not secreted in adulthood, which is common among Japanese people. Lactose ingested by drinking milk, for example, will proceed from the small intestine to the colon without being broken down. This results in a large buildup of lactose in the colon, which causes an increase in lactose-loving Bifidobacterium. However, a large proportion of people with European ancestry have the genotype that causes lactase to be secreted, which means that lactose is broken down and absorbed in the small intestine, and therefore less lactose reaches the colon. The inevitable result of this is less food for Bifidobacterium, and, consequently, an increase in the number of other microbes. Here, humans and microbes are in a competitive relationship concerning sugar, which means that the relationship between human genotypes and the quantity of microbes is maintained.
Conversely, there are cases in which humans and microbes have a cooperative relationship. The digestive enzyme amylase, which is found in saliva, is produced by the human salivary amylase gene, AMY1. The more of this enzyme an individual has, the more carbohydrates are broken down by saliva. Consequently, in people who secrete more amylase, complex sugars that are not broken down by saliva reach the colon, whereas in people who secrete little amylase, both complex and simple sugars reach the colon. This means that the quantity of Ruminococcus, which has a preference for complex sugars, increases in people who secrete more amylase. Accordingly, we know that the quantity of Ruminococcus can be predicted from looking at the AMY1 genotype.
We know that obtaining energy from lactose and carbohydrates became crucial to human survival as humankind developed from being hunter-gatherers to pursue agriculture and livestock farming instead. It has been suggested that not only the evolution of human genes, but also the accompanying evolution of gut microbes has contributed to human evolution. Partly because a number of such relationships between individual microbes and genes have become apparent in recent years, we thought there is a strong possibility that gut microbes codiversified with humans and embarked on this study.
Phylogenies can be constructed by finding DNA sequences
We first collected the saliva and feces of more than 1,200 humans from six countries across three continents: Africa (Gabon and Cameroon), Asia (Vietnam and South Korea), and Europe (Germany and the UK). We extracted DNA from these samples and used them to produce paired human genome and gut microbe metagenome datasets. We then created phylogenies representing their history of evolution and compared them against each other.
While techniques for creating phylogenies using human genomes and cultured bacteria have already been established, producing phylogenies for individual bacteria from a metagenome containing a mixture of all kinds of bacteria has been technically challenging. Due to the increased attention focused on gut microbes, various tools had already been developed by the time we started this study in 2018. Consequently, we were able to produce phylogenies by searching for specific DNA sequences called genetic markers specific to individual bacterial species.
Subsequently, we began by creating phylogenies for 58 bacterial strains and one archaeal strain commonly found in adults and examined them for codiversification with humans. Some microbial species showed strains specific to people in Africa, Europe, and Asia, as well as country specific patterns within those continents, emerged from the results (Figure 1). Statistically significant codiversification was observed in 36 of the 59 taxa. At the same time, strains completely unrelated to regions also appeared. Looking at these phylogenies, most of the bacteria demonstrating codiversification with humans originated in Africa, as we had anticipated.
- * Strain: cells derived from the same cell.
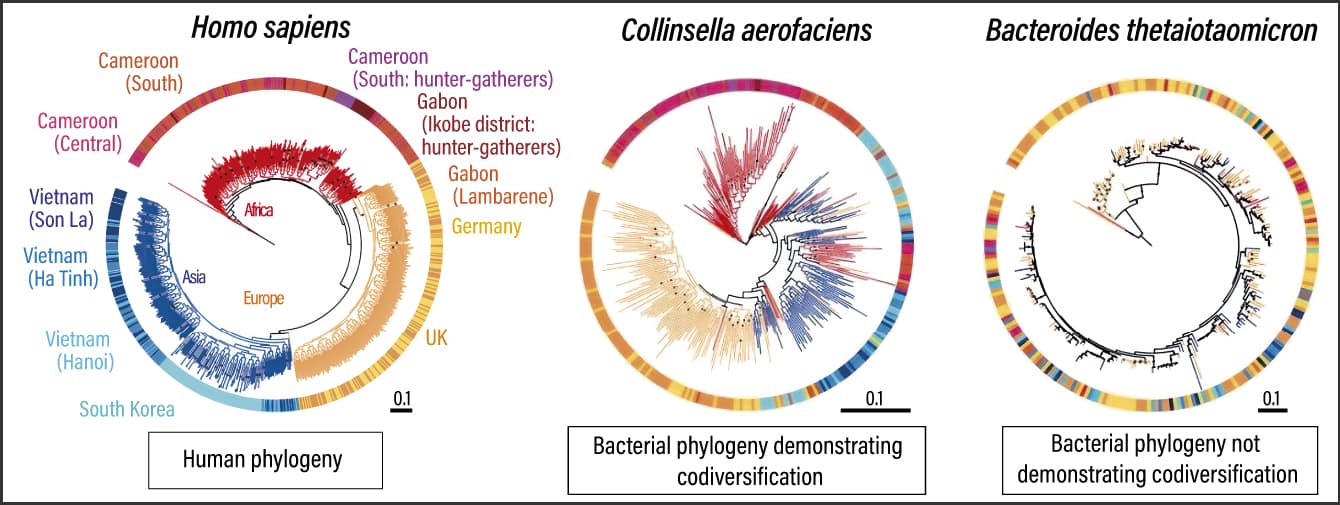
Figure 1. Codiversification of humans and gut microbesThe research team compared the phylogenies of humans and microorganisms, and measured the degree of codiversification. Bacteria with corresponding phylogenies —— that is to say, those that demonstrate codiversification —— have neatly grouped colors, whereas the colors are all mixed together in the case of those that do not demonstrate codiversification. Areas where they do not correspond are indicative of gut microbes having frequently migrated between populations. Some also indicate an intermediate phase.
In addition, when we conducted an analysis that also included published human metagenomes from 19 countries, metagenomes from 27 primate species, and metagenomes from the feces of humans who lived in North America 1,000-2,000 years ago, we further supported the findings mentioned above. For example, in the case of bacterial species that codiversified with humans, we obtained phylogenies indicating that strains specific to primates were the ancestors of the human strains. From previous studies by other teams that investigated codiversification of gorillas, chimpanzees, humans, and gut microbes, we know that primates and certain types of Bifidobacterium have been through the same evolutionary process, but our research was the first study to show codiversification of gut microbes between human populations. So, how do microbes that have codiversified with humans differ from those that have not?
Microbes with a higher degree of codiversification have a smaller genome
Given the nature of the gut environment and microbes, the results we obtained made perfect sense (Figure 2).
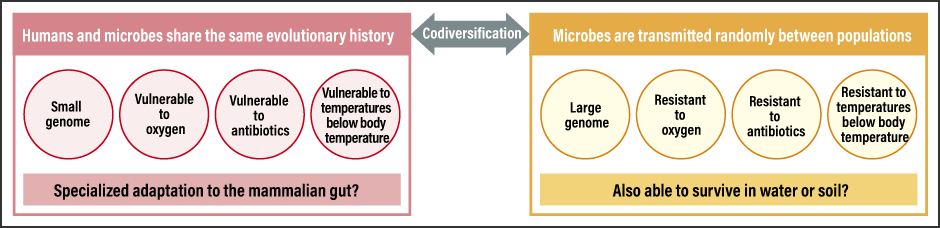
Figure 2. Gut microbe features dependent on presence or lack of codiversification with humansMicrobes that have adapted to the environment of the human gut exhibit unique characteristics suited to that environment.
First of all, anaerobic bacteria, which dislike oxygen, have clearer population-specific phylogenies. Because some gut microbes can live for a short time in water or air, their phylogenies do not show patterns indicative of codiversification with humans.
Another feature relates to temperature. When the microbes are cultured at body temperature (37°C) and at room temperature, codiversified microbes show poor growth in room temperature whereas those showing little codiversification are able to respond better to temperature changes, such as being at room temperature. Microbes demonstrating codiversification with humans prefer the stable temperature in the gut.
The third characteristic is antibiotic resistance. As microorganisms produce various antibiotics, microbes that live in the natural environment need to be resistant to them. Microbes therefore have numerous genes for responding to those substances. However, microbes that have undergone specialized adaptation to a habitat in the gut have little need for resistance to the antibiotics produced by other microbes, and consequently do not have the genes required to respond to them. As a result, they develop a lower resistance to antibiotics.
We also discovered that microbes with a higher degree of codiversification in the gut tend to have a smaller genome. Microbes in the natural environment need numerous genes to respond to that diverse environment, whereas microbes that have migrated to the stable gut environment do not require many genes. Maintaining a large number of genes comes with evolutionary costs for the microbe, as it slows down cell division and imposes the burden of gene maintenance for the purpose of propagation. Accordingly, we can predict that symbiotic bacteria have a smaller genome because they do not have any unnecessary genes. In the case of the insects and symbiotic bacteria mentioned at the beginning of this article, too, numerous examples of reduced genome size due to symbiosis have been confirmed, and there are even microbes that have virtually no genes at all. Human gut microbes are thought to be in the initial stage of this kind of symbiotic relationship. So clarifying the degree to which microbes that have codiversified with humans are dependent on us will be a topic for future study.
Right now, our team is keen to shed light on which microbes accompanied humans when they migrated, which microbes were left behind or replaced, how this was connected to human genes, and what impact it had on human evolution (Figure 3).
 ©Pixeleiderdown
©Pixeleiderdown
Figure 3. Human migration and gut microbe transmissionAs humans migrate across continents, microbes (strains) travel with them and progressively codiversified.
For example, there is a report on Helicobacter pylori (H. pylori) —— which can cause gastric cancer —— that is highly interesting from the perspectives of microbial evolution and human disease.
In South America, one Colombian town up in the Andes Mountains is known to have an incidence of gastric cancer that is high by global standards, at around 150 cases per 100,000 people. Many of the residents of this town belong to the indigenous population, and in residents who developed gastric cancer, the ancestral indigenous H. pylori strains had been replaced by European strains brought in by colonizers. On the other hand, residents of another Colombian town have an incidence rate of around 6 cases per 100,000 people. In the case of this second set of subjects, both the individuals’ genomes and the genomes of the H. pylori strains they carried could be traced back to African origins.
Having originated in Africa, H. pylori spread and codiversified as humans dispersed, with the result that humans gained population-specific strains of H. pylori. This study shows that individuals can coexist with the strain of H. pylori they originally had, and the pathogenicity differs when the H. pylori strain is replaced with strains with a different origin. To put it another way, a single microbial strain could coexist with some individuals, while causing others problems, depending on the human genotype. Another article makes a similar suggestion about the pathogenicity of the bacterium responsible for tuberculosis, Mycobacterium tuberculosis, which also originated in Africa and has codiversified with humans.
Differences in microbial species and strains based on genetic origins and regions
From our research, it has become clear that differences in microbial species and strains occur based on genetic origins and regions, even among human gut microbes. If so, does it mean that the same phenomenon seen in the case of H. pylori occurs among the gut microbes that have coexisted with humans since ancient times? And if true, how did this occur? We would like to trace the history of microbes that demonstrate codiversification with humans, to find out more about them.
I am currently based in the U.S. state of Arizona and conducting my research here. It goes without saying that the U.S. is a nation of immigrants. The people who have migrated here from a diverse array of countries likely carry microbial strains from the countries where they previously lived or were born. However, by the time several generations have passed since their ancestors settled, there is a possibility that people will have lost the original microbial strains their ancestors had, due to antibiotic use, dietary changes, and exposure to other microbial strains, for example.
In that case, we can investigate which microbes remain and which microbes or microbial strains are replaced after people migrate to a different place. We also believe that undertaking more in-depth research into human genes and the microbes that humans maintain across the generations could help unraveling the evolutionary process of how humans have adapted to their environment.
At the same time, we hope to explore how coevolution suddenly breaks down due to mismatches between human genome and the microbial genomes, and examine the consequence on human traits and disease.
The previously mentioned H. pylori example is also thought-provoking from the perspective of shedding light on the causes of disease. In that example, individuals with African genomes are not harmed by microbial strains with African ancestry, but in non-African individuals, the presence of African strains may trigger the onset of cancer. In other words, microbial strains are interacting with some kind of genes in the host, and show pathogenicity if these host genes are not absent.
As gut microbes are one element with a substantial impact on human health, particularly immunity, we believe looking at this relationship will also lead to disease prevention in humans. Today, the relationships between genes and various diseases have become clear, and genetic testing has made it possible to provide statistical estimates of the probability of disease in individuals. It is likely that taking information about microbial genes and the codiversification of gut microbes into account will lead to disease prevention and treatment in the future. For example, we may be able to inform patients, “With your genotype and ancestry, you should have these strains of these microbes, but you actually have strains from another region.” This information could potentially be combined with genetic testing so that your gut microbes could fulfill a role akin to a kind of biomarker. We are already embarking on the first step by investigating so-called “modern diseases” closely related to immunity, namely allergies and asthma.