It would be fair to say that the invention of the microscope marked the beginning of modern biology. Later on, the groundbreaking discovery of DNA’s double helix structure established the field of molecular biology. It was hoped that recent successes in analyzing genome sequences would bring about a further leap forward in our exploration of life, but in fact, the mystery has only deepened. In other words, genetic information alone is not sufficient to understand how the proteins produced act and how they are configured. To find out the principles of life itself, we need to actually look at their activity; and to do this, live imaging is essential.
Special Feature 1 – The Evolution in Visualization Intracellular visualization will help us zero in on the principles of life!
composition by Rie Iizuka
Biology originated with the act of looking at things. It was back in the latter half of the 17th century that Dutch textile merchant Antonie van Leeuwenhoek invented a single-lens microscope. Using this microscope, Leeuwenhoek discovered the existence of microorganisms and sperm. At around the same time, English scientist Robert Hooke, who was a member of the Royal Society, succeeded in observing the cell walls in a cork with a microscope. As these cell walls resembled honeycomb cells, he named them “cells.” This is the origin of the biological term “cell.”
Genome analysis is not enough to understand life
The discovery of X-rays and electrons at the end of the 19th century was followed in the early 20th century by the start of efforts to develop electron lenses. The development of the electron microscope in the early 1930s dramatically increased resolution, making it possible to carry out observation at the molecular level.
In tandem with instrument development, the field of biology achieved remarkable progress in 1953 when the double helix structure of DNA was posited. Molecular biology became established as an academic discipline and many studies have yielded positive outcomes since then.
Cutting-edge technology is behind the advances in these academic fields. The development of groundbreaking techniques such as DNA sequencing in the latter half of the 1970s and polymerase chain reaction (PCR) in the 1980s has brought new perspectives to bear on the realm of biology.
2003 saw the declaration that the human genome had almost been decoded. Later, in 2022, an international team including scientists from the U.S. National Human Genome Research Institute announced that they had completed the remaining 8% of the genome, which had been difficult to sequence using the techniques available at the time.
The whole world was thrilled by the thought that unraveling the genome sequences of humans and numerous other organisms would swiftly shed light on the mysteries of biological phenomena. However, in fact, genome analysis alone turned out to be totally insufficient for us to understand life and we found that further research would be required.
Obtaining genetic information by analyzing the base sequences of organisms gave us a comprehensive set of information about proteins, which are a key component of the body. Nevertheless, that on its own still leaves us a long way from having unraveled the mystery of life.
This is because, even though we have information about proteins, we do not know what circuits these components produce or the timing of the introduction of each component, along with where and how this takes place.
For example, if we think in terms of a plastic model kit, the information about the parts in the kit should be enough to enable us to build the model. However, while we know the components that make up an organism, we still cannot make that organism from scratch.
Visualizing the reactions occurring inside cells
One reason why we cannot make an organism is that we do not know how the components work, in terms of how the circuits of components are connected, and when and where those parts are incorporated. If understanding life means we can build it from components, knowing the sequences of those parts is not enough to allow us to reach that goal; we still need to be able to see the situation within the circuits in which those components function.
In other words, to understand life, we need to try to track the reactions occurring within living cells in real time, visualizing the activity inside them and recording it just as it occurs.
The technique that has emerged for this purpose is the one on which I work: live imaging.
This technique originated with the discovery of green fluorescent protein (GFP). Dr. Osamu Shimomura, who was awarded the Nobel Prize in Chemistry, achieved a world first in the 1960s when he discovered GFP in the jellyfish Aequorea victoria. The protein he discovered emits green fluorescence when exposed to blue light. Due to the identification of the gene sequence required to synthesize GFP, scientists developed a technique for introducing that sequence into various cells and using GFP as a marker for visualizing the subject (Figure 1).
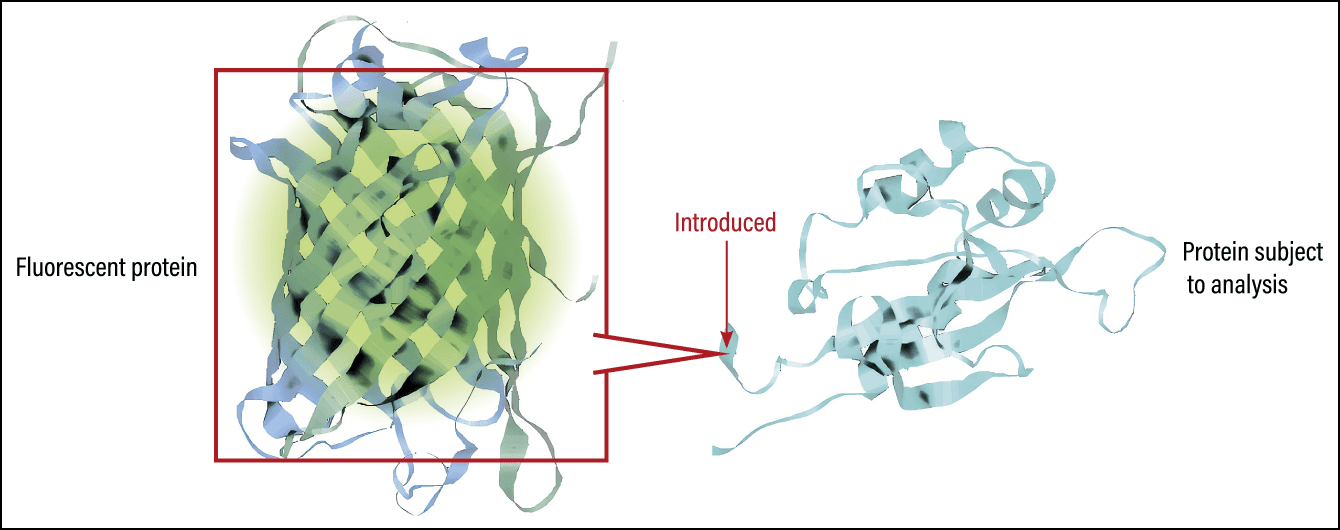
Figure 1. Illustration of the use of fluorescent protein for visualizationIntroducing a fluorescence-emitting protein such as GFP into a cell makes it possible to track that cell.
Detecting the introduced GFP is simple, as it merely involves exposing the cells to a specific type of light, and this technique has become so prevalent in biological research that it would be fair to say that everyone working in this field has used it.
Aside from GFP, there are other fluorescent proteins that serve as outstanding tools. As they do not require a separate substrate and do not exhibit cytotoxicity, they have hardly any effect at all on the target cells. In addition to that initially obtained from Aequorea victoria, a fluorescent protein from amphioxus also demonstrates a high level of stability, while fluorescent proteins derived from coral and sea anemones can express colors other than green, thereby expanding the range of colors that can be used to stain cells. The availability of a more abundant array of colors means that if one wants to observe the interaction of a nucleus with two different molecules, A and B, one can stain the nucleus blue, molecule A red, and molecule B yellow, for example. One can then observe their position to see whether the molecules are located inside or outside the nucleus, or watch them entering and leaving the nucleus over a period of time. Thus, the age has arrived when we can observe the interior of a cell in color.
Omics —— the collective term for the biological disciplines that involve comprehensively investigating the molecules within an organism, by such means as genome analysis —— specializes in analyzing the average condition of a large number of cells in a stable state. It is thanks to omics that we have identified base sequences, and this domain is making a particularly significant contribution to the field of medical care. As a result, we are now able to deliver high-precision medical care, including the ability to select the appropriate drug in light of an individual’s genomic information.
Seeking to learn about the place where life actually happens
On the other hand, the important thing about live imaging technology is the ability to view the situation as time passes. As omics provides a snapshot of a moment in the life of a cell, it cannot show us how the cell behaves in the next moment. Furthermore, there are individual differences between cells, so not all of them necessarily behave in the same way. We seek to use imaging to track information in individual cells, such as fluctuations in the number of molecules inside them and changes of stage, in order to learn about the place where life actually happens.
One biological phenomenon that we have visualized using our technique is the behavior of a cellular slime mold in a state of starvation. Described as a social amoeba, Dictyostelium discoideum is a eukaryote found in soil.
Figure 2 shows the slime mold in a state of starvation. The nuclei are stained red. Slime molds usually use bacteria in soil as their source of nutrition, but when they are in a state of starvation due to a lack of such bacteria, these slime molds exhibit highly interesting behavior. Hundreds of thousands of cells flock to the center and, over the course of around 16 hours, form a slug-like shape. They then seek out a good place where they can create their ultimate structure, fruiting bodies, for the purpose of producing spores and becoming dormant.
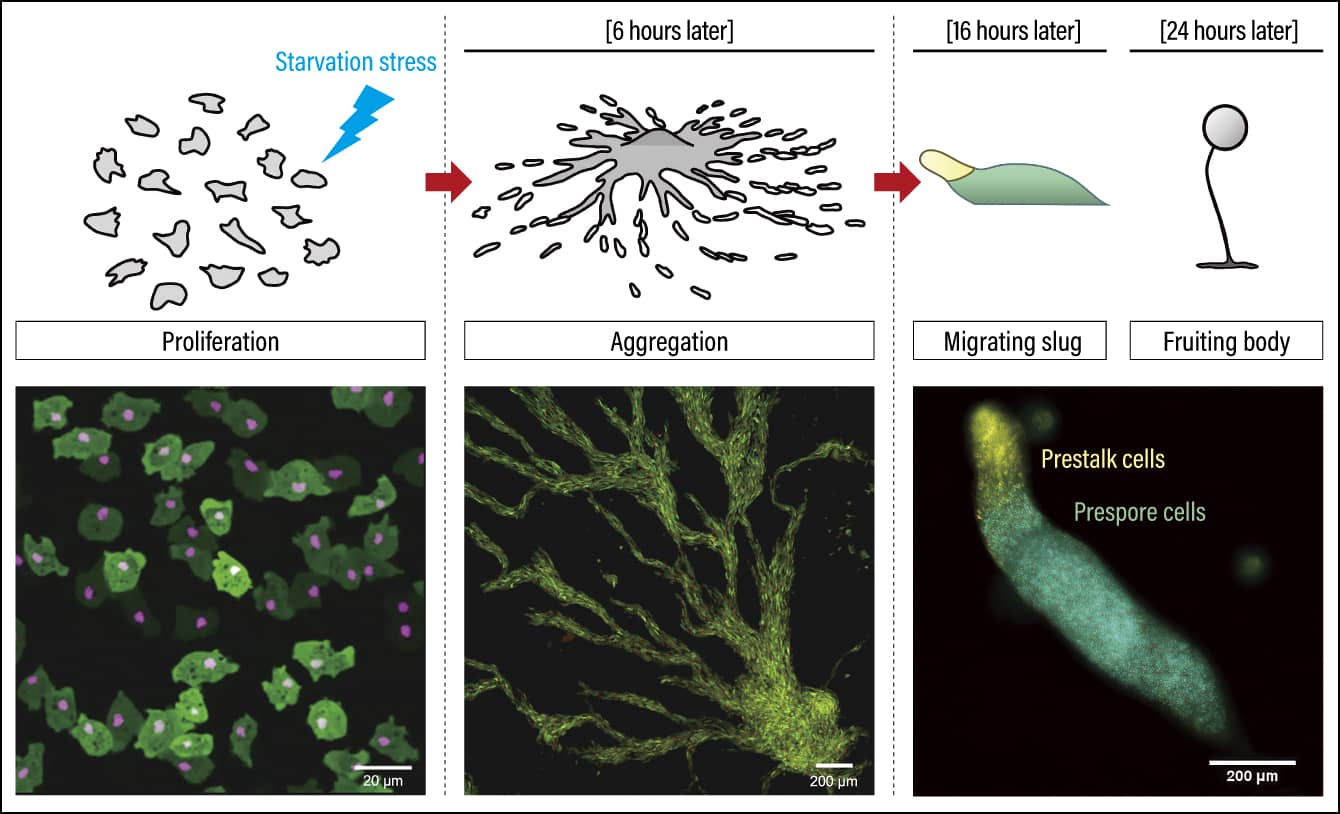
Figure 2. Behavior of Dictyostelium discoideum in a state of starvation (sequence photographs)Each of the scattered red spots is the nucleus of a slime mold. These gradually aggregate in a single location and eventually form a slug-like shape.
Initially, one cell starts to move, then the other cells follow suit, gradually swirling toward the center; once they have all aggregated there, they form a shape, which then moves to another place.
Just being able to observe this series of movements is enough to make one exclaim in wonder, but for those of us who work in a biology laboratory, it also raises questions about how this puzzling phenomenon of amoeboid cells developing the ability to engage in collective cell migration could occur in the first place.
This behavior in which the cells aggregate involves cell adhesion. Accordingly, we sought to shed light on the mechanism by observing the transcription (the process in which RNA is synthesized in accordance with the DNA base sequence) of molecules that promote cell adhesion (Figure 3).
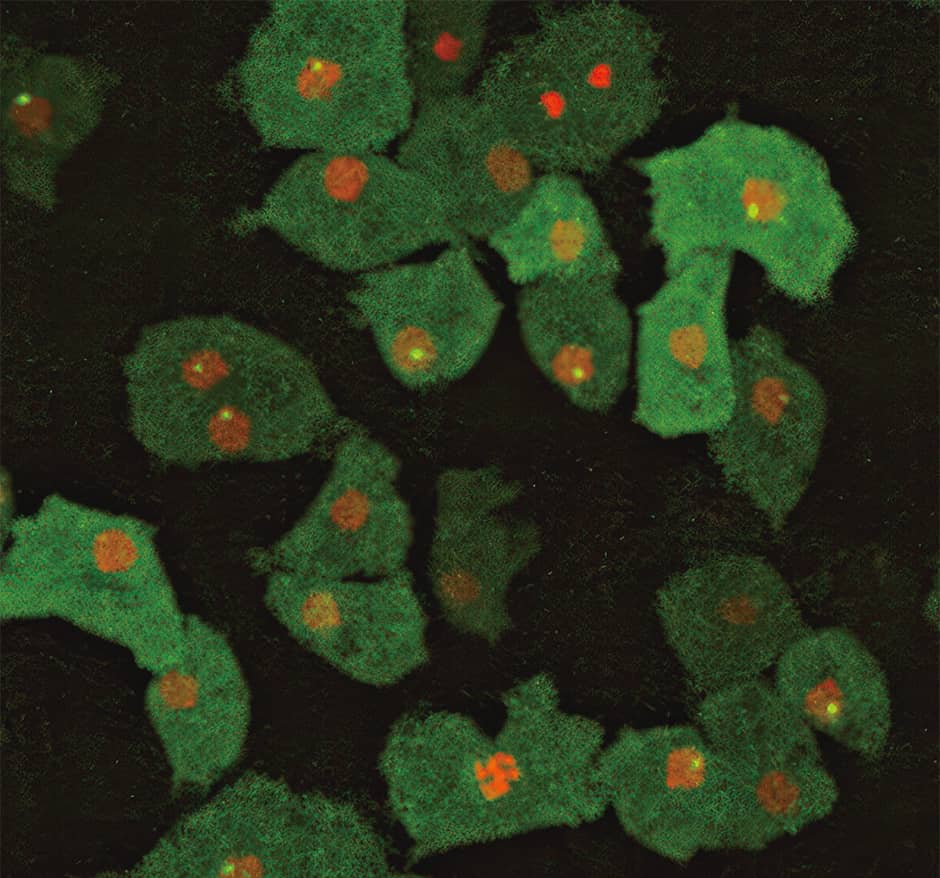
Figure 3. mRNA transcription in progressBright spots visible in the nuclei indicates that transcription is taking place. When no bright spots are visible, transcription is not taking place. The transcription situation varies from one cell to another, with the spots showing up brighter when a lot of mRNA is being produced, and darker where there is little mRNA.
In the figure, bright spots can be seen in the nuclei indicated in red. These are the visual representation of the moment when the enzyme that synthesizes messenger RNA (mRNA) from DNA starts to work. The fluorescent molecules do not adhere to nucleic acid —— DNA and mRNA —— so it does not light up on its own. We visualized the moment when mRNA is produced as a bright spot in the cell. This was achieved by binding a special protein fused with a fluorescent protein to mRNA required to synthesize molecules for cell adhesion. Looking at a video, we were able to observe the process of transcription being repeatedly turned on and off, with the bright spots suddenly appearing at the moment of mRNA transcription before disappearing again.
Binding the fluorescent protein to mRNA and tracking the transcription spots is quite a time-consuming experiment. Nevertheless, we developed this process because we believed that if we were to discuss quantitative fluctuations in mRNA in fundamental terms, it was essential to observe mRNA directly, rather than looking at a synthesized protein.
What happens inside cells?
mRNA is synthesized when a cell is trying to perform some kind of action. To illustrate it in anthropomorphic terms, one could say that a cell that has observed its state of starvation issues the order to assemble and move; the cells around it aggregate in response to its “voice” and the circle of aggregation spreads out. Once a certain number of cells have aggregated, they line up in the appropriate formation and start moving elsewhere.
Looking at a video capturing the molecules that control mRNA synthesis, we can see that they start to activate in the process of the cells aggregating, and that the molecules oscillate. We also discovered that the collective motion is synchronized with this oscillation.
In addition, we succeeded in controlling the behavior of slime mold in a state of starvation by manipulating the transcription of these molecules.
In other words, by looking at their actual movement, we were able to gain a partial understanding of what is occurring inside cells and why unicellular organisms can engage in synchronized collective motion as though they have a brain.
One can only marvel at the mystery of life when one actually watches slime molds, which do not have brains, respond to a crisis in the form of starvation by engaging in collective motion as if following prescribed rules.
We carried out research into the visualization of transcription because we wanted to view the phenomenon of what is known as the central dogma, which states that the genetic information stored in DNA prescribes the structure and function of proteins via RNA. Another outcome of our study was the finding that, rather than mRNA transcription being constantly on, the chemical reaction cycle differs from one cell to another and even within a single cell.
For example, rather than being homogeneous, as had previously been thought to be the case, cancer cells are actually heterogeneous. Looking at molecule transcription in progress in our research, one can certainly see that it is not uniform, and that there is clear variation in the frequency of gene expression even within a cell with the same genes, as can be seen in Figure 3.
Observing such activity in the unicellular world expands our thinking in a diverse array of directions. If we liken it to an ecosystem, the interaction between cells could be viewed as akin to the interaction of individuals at various levels. In response to information from one individual, others face the same direction and, before long, the entire group starts to move at an energetic pace. Similar things frequently occur in human society, too. It feels as though one has observed a kind of fundamental principle of biological organisms.
My own desire to see the workings of life was triggered by the realization that genome analysis alone does not provide us with enough knowledge to understand life, but a background factor was probably my very personal experience of observing organisms almost daily along the banks of a river near my home when I was a child. As I watched a crab waving its pincers around, I was struck by a very simple question: why do they do that? This is the basis of my research, and I think it is the fundamental question underlying the entire discipline of biology.