Genome editing is a technology that involves modifying DNA by cutting base sequences and joining them to other sequences. Genetic modification is a form of genome editing. More advanced methods are now emerging and are being commercialized in a variety of fields, including medical care and energy, not to mention foods. At the same time, precisely because the modification of genes has a direct impact on life, cautious discussion of safety and bioethics involving a broad swath of society is of paramount importance.
Special Feature 1 – An Introduction to Genome Editing Advances in editing technology will make modification safer and more efficient!
composition by Rie Iizuka
The word “genome” refers to the full set of genetic information contained in an organism’s DNA. As genetic information determines an organism’s characteristics, “editing a genome” means altering an organism’s characteristics by modifying its genes. The CRISPR-Cas9 genome editing technology was developed in 2012, since when its use has become prevalent in a variety of fields.
First of all, let us look at what modifying genes actually involves.
Genes are the blueprint of life, consisting of bases called adenine (A), thymine (T), guanine (G), and cytosine (C). Gene modification begins by cutting into this genetic blueprint somewhere along its length. Then, DNA’s own repair mechanism is used to rejoin the cut gene or introduce another sequence. This gene modification technology was first established in 1973—more than 30 years before what we know of today as genome editing first appeared on the scene (2005)—in the form of a method that involved severing the double strand of DNA using a restriction enzyme and repairing it in bacteria.
Success in snipping solely the selected sequence
So how does genome editing technology differ from conventional modification technology?
The most notable features of genome editing are its efficiency and its specificity. To start with, it enables the DNA in the target area to be snipped—the first step in modification—with a high level of precision.
“Snipping the DNA” means causing a chemical reaction that unfastens the bonds between the bases and enzymes called nucleases, which are present in all organisms, had been used to do this since modification technology was in its infancy. There are a number of types of nuclease, including restriction enzymes that sever only DNA, those which snip only RNA, and those which cut only a specific sequence. In the latter half of the 1990s, scientists created artificial enzymes called zinc finger nucleases (ZFNs) and succeeded in snipping only the sequences that they had selected. This technique is still widely used in clinical practice today.
The next technology to be created was transcription activator-like effector nucleases (TALEN), which were developed from pathogenic bacteria from plants. It was particularly useful, because TALEN recognize only a single base.
And then, in 2012, CRISPR-Cas9 was developed. It is widely used today because it is easy to use and highly efficient.
CRISPR-Cas stands for Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR associated protein. While this term might generally call to mind a genome editing tool, researchers who study microorganisms know it as the immune system of bacteria and archaebacteria (Figure 1).
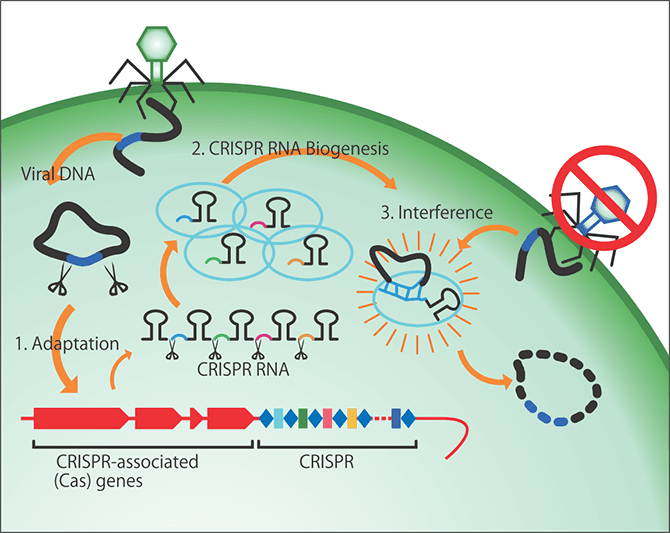
Figure 1. CRISPR-CasDiscovered in the form of the acquired immunity of bacteria and archaebacteria, it was later developed into a genome editing tool. Archaebacteria incorporate fragments of exogenous DNA from phages that infect them and sever the exogenous DNA by synthesizing homologous RNA. The areas indicated by the light blue ovals incorporate the phage and then Cas9 and guide RNA (gRNA) leading to the target DNA sequence work together to sever the targeted gene.
The CRISPR-Cas system in bacteria behaves just like acquired immunity in humans. Viruses that infect microorganisms are called phages. When infected by a phage, a bacterium incorporates it and remembers it, so the next time that bacterium is infected by the phage, it specifically destroys that phage. The discovery of this mechanism in bacteria had a huge impact and an American researcher went on to use the CRISPR-Cas system to develop genome editing technology.
These genome editing tools are excellent systems capable of cutting specifically targeted areas of DNA, which, in humans, consists of 3 billion base pairs. It is this property that makes genome editing so revolutionary. But why is it able to cut specifically targeted areas when this had previously been so difficult?
Able to transcribe even a single base
Let us take humans as an example. Looking at the sequence pattern of the bases A, T, G, and C, the probability of the four letters being aligned in the same sequence is 4 × 4 × 4 × 4 = 1 in 256 letters. In other words, if we think in terms of four-letter units, the same sequence will appear once every 256 letters. The restriction enzymes used before genome editing emerged broadly recognize 4-8 letters at a time, so if the same four-letter sequence occurs once every 256 letters, there is a possibility that an eight-letter sequence will appear once every 65,536 letters. As there are 3 billion base pairs in human DNA, scientists had to snip the target DNA multiple times and wait until they got “lucky.”
However, CRISPR-Cas9 recognizes strings of 20 letters. Four to the power of 16 works out at more than 4 billion, so a pattern of letters that is 20 letters long (4 to the power of 20) would only be found once in 3 billion pairs. In other words, as there is a high probability that a 20-letter sequence will only appear once in one specific location in the 3 billion base pairs of DNA, it is possible to transcribe one specific targeted DNA sequence and even a single base (Figure 2).
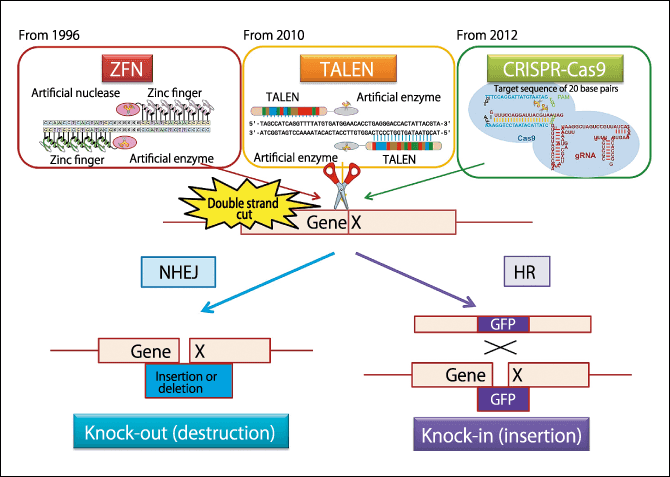
Figure 2. Genome editing using ZFN/TALEN/CRISPR-Cas9The artificial enzyme severs the gene at the target location. As it can make gRNA capable of binding to the targeted 20-letter sequence, it recognizes the specific sequence, enabling CRISPR-Cas9 to be used to make the snip. We can now make “scissors” that cut only the places we want to cut, while leaving untouched the places we do not want to cut. Either the DNA is joined up as it is (deletion) or another gene is incorporated (insertion). Then, using homologous recombination (HR), a Green Fluorescent Protein (GFP) gene, for example, is inserted into the severed portion, enabling the gene to be traced thereafter.
There are two methods of repairing DNA. One is called non-homologous end joining (NHEJ) and, in simple terms, involves joining the ends of snipped DNA. While the DNA sometimes reverts to the original sequence immediately after snipping, the nature of some cells is such that it sometimes results in deletion of part of the DNA and removal of the sequence that previously existed. In genome editing, this is called knocking-out a gene, which means deleting a gene that has a particular function.
The other repair method is called homologous recombination (HR).
Humankind has a pair of chromosomes from parents. This means that even if the DNA within a cell is cut, the similar sequence in the other chromosome can be used like a template to repair it. This is the method called homologous recombination. In genome editing, recombination is brought about by inserting the desired sequence into the disconnected part of the DNA. This is called knocking-in.
No genes are introduced from outside
People often ask how genome editing differs from genetic modification. One could describe genome editing as the high-level conceptual version of modification as it existed previously.
Let us look at an example. In a pufferfish created by genome editing to have more flesh than ordinary pufferfish, just one of its 10,000 genes—the gene called myostatin, which is related to muscle composition—has been destroyed and no genes have been introduced from outside the fish (exogenous genes). This is a mutation that is perfectly capable of occurring in the natural world in pufferfish of this kind, without the need to use genome editing technology. Genome editing simply increased the probability of its occurrence.
In addition, there has been what is called “selective breeding,” in which radiation or special chemicals were sometimes used to destroy a gene. However, that can cause the destruction of or unintended mutations in genes other than that targeted, or even the death of the cells themselves. In this sense, it is fair to say that genome editing and other such technologies that modify only a specifically targeted area are more precise.
On the other hand, genetic modification sometimes involves introducing into the disconnected portion of DNA a gene that the organism did not originally have. This is a mutation that cannot occur in the natural world. While genome editing can be used to modify DNA in this way, it must be understood that not all genome editing involves introducing exogenous genes.
At the same time, Japan has strict limits on the use of genome editing, with researchers obliged to notify the authorities and seek approval for using genome editing to create produce; they are also required to seek permission to use such produce in experiments.
Japan has also ratified the Cartagena Protocol, an international agreement on the handling of genetically modified organisms and organisms whose genes have been modified are subject to rigorous control to ensure that they do not proliferate and become incorporated into other organisms.
Regarding the labeling of items that have undergone genome editing, the Ministry of Health, Labour and Welfare currently states that it must be notified of organisms into which exogenous genes have been introduced, but there is no labeling requirement for items in which genes have been destroyed, because they are virtually indistinguishable from conventionally bred organisms that are the result of repeated interbreeding. Future decisions on labeling will, in practice, likely take into account consumer attitudes.
In addition, therapies that use genome editing are also garnering attention.
Genome editing therapies can be broadly divided into two types: in vitro therapies that involve removing the cells from the body to carry out genome editing before returning them to the body, and in vivo therapies, which involve triggering mutations by introducing CRISPR-Cas into the body (Figure 3). The former is familiar from its use to treat diseases of blood cells and immune cells, as well as in the treatment of gene-related diseases by means of induced pluripotent stem cells (iPS cells). As it can be applied once the safety of the genome-edited cells has been confirmed, it is considered the safer method and is therefore currently progressing toward practical application. In 2017, there were reports that genome editing and chimeric antigen receptor T cell (CAR T-cell) therapy had been used for the first time ever to treat a baby with leukemia. However, as there are diseases that cannot be treated solely via in vitro methods, clinical trials of in vivo genome editing are beginning around the world.
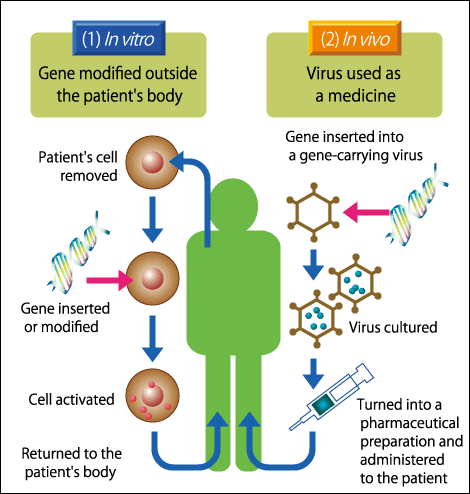
Figure 3. Therapeutic genome editingWhen considering genome editing therapies, we need to divide them into two categories: those carried out outside the body and those carried out inside it. The former is steadily progressing toward practical application. The latter type of treatment is irreversible, so greater safety needs to be established.
Thus, genome editing is an innovative and useful tool. However, we researchers must provide easily understood explanations of how these technologies are being used. In addition, we must develop even safer technologies. I believe that cautious discussion of the safety and ethics of modifying genes that involves society as a whole is particularly important.
Developing a rapid diagnostic kit for COVID-19 in Japan
Unfortunately, Japan is lagging behind the rest of the world in scientific research using genome editing. One reason for this is that the key genome editing tool CRISPR-Cas9 cannot be used easily, because it was developed in the U.S.A. and a huge royalty has to be paid by those using it. Naturally, CRISPR-Cas9 in its current form is not a perfect tool and other countries are frantically trying to develop their own, safer tools that they can use freely.
My team has developed a new tool called CRISPR-Cas3, which we have made available for use by Japanese companies. CRISPR-Cas9 uses the immune system of Streptococcus, but a number of other bacteria also have the same immune system. CRISPR-Cas3 uses the mechanism of a different bacterium and is capable of recognizing 27 letters, so we believe it is even safer than CRISPR-Cas9.
CRISPR-Cas3 and other such tools serve as “scissors” in genome editing. We have used this function to develop a kit for rapid diagnosis of COVID-19 using a method called Cas3 operated nucleic acid detection (CONAN) (Figure 4).
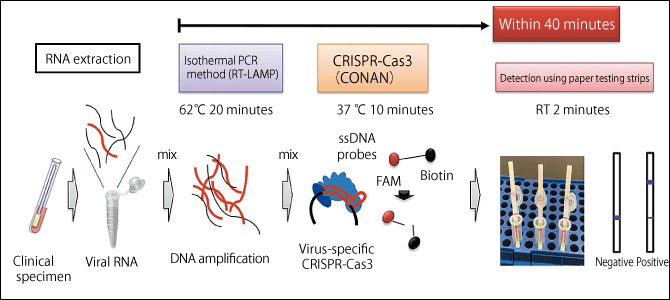
Figure 4. Overview of COVID-19 detection using the CONAN methodThis method is capable of detecting the virus in as little as under 40 minutes after taking the patient’s sample. As it requires only ordinary reagents, paper testing strips, and a device to keep the samples warm, it is quick, easy, and cheap. CRISPR-Cas3 can also detect changes in a single base, so a detection method can be established immediately if the virus mutates in a way likely to lead to drug resistance or a more severe form of the disease.
There are currently three tests for COVID-19: antigen, polymerase chain reaction (PCR), and antibody. The antibody test examines the number of IgG and IgM antibodies after infection, but this only shows whether that person has contracted the disease in the past and does not indicate whether or not they are currently infected. PCR testing or antigen testing are the only means of ascertaining the current situation and preventing the spread of infection, but while PCR tests are the more sensitive method, they take a lot of time and money. Antigen tests, on the other hand, are a simple test capable of finding the virus surface protein, but they have disadvantages in that they do not detect the virus where only small amounts of viral protein are present and can end up detecting similar viruses. Accordingly, we have overcome these two disadvantages by using CRISPR-Cas3 to develop the CONAN method for antigen detection.
The CONAN method recognizes the COVID-19 virus and then simultaneously severs surrounding markers and makes them light up. We have successfully used it to detect the virus accurately in as little as under 40 minutes. Thus, as seen from these examples of the use of CRISPR-Cas, genome editing has become an essential tool that is widely used everywhere from basic to applied research in a range of fields, including medical care, agricultural produce, and energy.