Insects account for as many as two-thirds of all creatures on Earth. Most of them have wings, but we do not actually know where the evolutionary origins of those wings lie. This debate got underway as early as the latter half of the 19th century, when On the Origin of Species was published, but to this day, the puzzle has never been solved. However, research using genome editing means that we might finally see the day when the answer to this conundrum is revealed.
Special Feature 1 – An Introduction to Genome Editing Has science finally solved the long-standing puzzle of where insect wings came from?
composition by Rie Iizuka
Insects first appeared on land between 400 and 500 million years ago and subsequently acquired wings. Earth’s named species of insects alone number in excess of 1 million and it is said that gaining wings was what enabled insects to thrive and proliferate to this degree.
However, the evolutionary origins of those wings have long remained a mystery. This is because there are no fossils that serve as the insect equivalent of Archaeopteryx for birds, so scientists have been unable to track their evolution.
Among insects that currently exist, the firebrat (Figure 1), for example, is a close phylogenetic relative of pterygotes (winged insects), but does not develop wings upon reaching adulthood and spends its whole life as an apterygote (wingless insect). The pterygote group includes some of the oldest species, such as dragonflies and mayflies, which zoom around on their splendid wings. However, when comparing firebrats and dragonflies, scientists have been unable to find any kind of process or anything else suggesting vestiges of wings.

Figure 1. FirebratThis species retains a primitive form of an insect that does not have wings.
The epicoxal hypothesis and the paranotal hypothesis
Debate over insect wings began soon after On the Origin of Species was published in the latter half of the 19th century.
The epicoxal hypothesis and the paranotal hypothesis emerged from the discussion and have long been regarded as the most likely explanations (Figure 2). Mayfly nymphs and other immature aquatic insects have paddle-like gills in their abdomens. As larger gills provide greater respiratory efficiency, it is only natural that these organs would evolve to become bigger. Gills closely resemble wings in shape and the theory that they evolved into wings is known as the epicoxal hypothesis. The epicoxal hypothesis is still an integral part of the debate today.
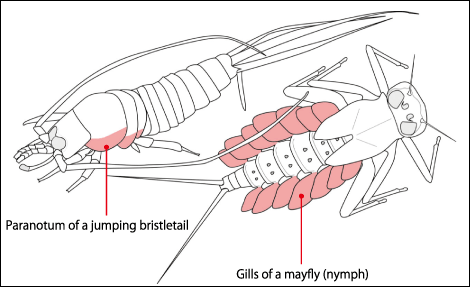
Figure 2. The two main hypotheses concerning insect wingsThere are two theories concerning the origins of insect wings: the epicoxal hypothesis, which holds that they developed from gills in insects’ aquatic ancestors, and the paranotal hypothesis that they are enlarged paranota.
On the other hand, the paranotal hypothesis emerged because the wings in hemimetabolous insects are formed from the paranotum. This theory holds that, at some stage in the evolutionary process, while insects would tilt their bodies to glide or minimize the impact on landing when jumping or when falling from a high place, the paranotum increased in size and gained joints, turning into wings with which the insect could fly.
Both theories refer to structures that are plausible as the origins of wings. In the case of the epicoxal hypothesis, it is mayfly gills as well as the epipods in crustaceans, which are the ancestors of insects, while the paranotal hypothesis focuses on the paranotum formed in apterygote insects such as firebrats and jumping bristletails. There are debates about each individual species, but every species can broadly be classified as falling into one or other category. In addition, as there are no species or fossils to indicate the stage at which wings developed, the 20th century saw ongoing discussion about which of two elements that are key features of wings developed first: the lobes or the joints.
What broke this stalemate was genes. The addition of the genome concept to the debate in the mid-1990s enabled proponents of both hypotheses to think about what kind of genes were involved in wings and how those genes function to create the structures in which wings had their origin.
The analysis of fruit fly (Drosophila) genes is very well advanced and most of the genes involved in wing formation have been identified. Once it became clear that wingless insects also had those genes, scientists began to investigate what the genes related to wing formation did in wingless insects and how those genes were used in the structures in which they originated in each insect species. Something akin to “comparative embryology 2.0,” the field of evolutionary developmental biology emerged once we were able to add genetic information to conventional information about form and development in discussing differences in shape between species.
Proof confirming the sites where the “wing genes” are expressed
Teams using the evolutionary developmental biology approach to study the origins of wings emerged between the late 1990s and the early 2000s. Then, in around 2010, a team from Japan’s RIKEN, the Institute of Physical and Chemical Research, succeeded in finding where the “wing genes” are expressed in jumping bristletails and mayflies.
Jumping bristletails are apterygote insects which retain features from before wings evolved and have paranota on their thoracic segments. On the other hand, mayflies have gill structures, as described above, and it has been reported that the wing genes are expressed in both gills and paranota. In other words, the origin of wings lies not in one or the other, but rather in both structures. While these genes are just some of the genes involved in the development of wings in fruit flies, the discovery showed that they are expressed in both these structures, which had until then been regarded as totally different, and therefore brought science a lot closer to a combined hypothesis bringing together the epicoxal hypothesis and the paranotal hypothesis.
While this was in itself a big step toward unraveling the mystery, it only showed that the genes were expressed in those parts. To investigate how those genes actually work and what interaction they have with other genes, it is necessary to conduct analyses that involve eliminating the wing genes and expressing them in other places. A single gene does not necessarily have only one function, while the same gene might have different functions in different species. Even within a single individual, genes are frequently expressed in different places during an organism’s life cycle; for example, a gene might be expressed in the back at one point and in the legs at another. That is why it is hard to say that the wing genes are responsible for the formation of wings just because they were expressed. Accordingly, genome editing is going to take on an important role.
Analysis made possible by genome editing
When the field of evolutionary developmental biology gained a new tool in the form of genome editing, it became possible to undertake detailed analysis by destroying genes or inserting them into all kinds of organisms. This enabled scientists to track the fate of a specific cell in a living creature, for example.
In my own research into the origins of wings, I compare crickets and firebrats. Adult crickets have wings, but the nymphs closely resemble apterygote nymphs when they first hatch. The only difference is whether or not they develop wings after molting, so I believe that comparing the two species will make it possible to find out why apterygote insects does not make wings.
As described above, a great deal of research into the genes required to make wings has been carried out in fruit flies, which are model organisms. However, they are holometabolous insects and one could go so far as to say that the cells which form the larvae and the adult fruit flies are completely different, which makes it unrealistic to compare them directly with apterygote insects, whose form hardly differs at all between the larval and adult stages. Previously, it was not possible to destroy a particular gene to investigate its role in a non-model organism, such as crickets or firebrats.
However, the emergence of genome editing now enables us to do just that with all organisms. I myself reported success in editing the genome of the firebrat Thermobia domestica in 2018. I am currently using genome editing technology to investigate the ancestral roles of wing genes in crickets and firebrats by modifying a gene that is believed to be related to wing formation in fruit flies.
Around the world, some teams are analyzing the role of the same gene in mayflies—which form gills—in an effort to verify the epicoxal hypothesis, while others are analyzing crustaceans, which are the ancestors of insects. Thus, thanks to the emergence of genome editing, we are now able to conduct detailed research using a variety of organisms and I have the impression that tests focused on this 160-year-old mystery are progressing rapidly.
Another important example of the use of genome editing is the technique of inserting a reporter gene such as green fluorescent protein (GFP). For example, if you insert it close to a gene considered to be involved in wing formation, it will make the cells in which the gene is expressed visible, enabling researchers to examine living organisms to track whether or not those cells actually form wings.
This technique has also led to the subsequent development of the aforementioned combined hypothesis. After a Japanese research team reported that both gills and paranota have structures in which wings originate, a U.S. team published an article stating that, in a species called the red flour beetle (Tribolium castaneum), two groups of cells originated in different places merged and formed wings after going through some kind of process. While the combined hypothesis is also supported by studies like this and it is a widely held theory explaining the origins of wings, I feel that it lacks conclusive evidence. This is because nobody has yet observed the process of the cells merging to form wings. However, if we could use genome editing to separately label the relevant cell groups, we would likely be able to directly prove the hypothesis as to whether or not the cells actually move and merge. Alternatively, if apterygote insects also have equivalent cell groups, what might their fate be? Genome editing has made experimental approaches like these possible.
And that is not all. A number of elements influence each other to determine the behavior of genes, such as the region of DNA called a promoter, which regulates gene activity, and the region of DNA called an enhancer, which adjusts the strength of that activity (Figure 3). When it comes to genes, attention tends to focus on the sequences that encode proteins, but they themselves do not vary a great deal. In fact, there have been many reports of cases in which evolution of an organism’s shape occurs when the DNA sequences of these enhancers and promoters—the regions regulating gene behavior—change. For example, regarding fruit fly patterns, we already know that the genes themselves are the same and that the places where the patterns emerge differ according to mutations in the regions controlling their expression.
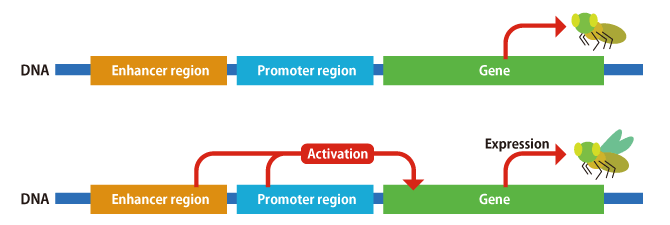
Figure 3. Gene activity/transcriptionDNA contains not only genes that encode proteins, but also promoters, which regulate the activity of genes, and enhancers, which adjust the strength of a promoter’s effect. As some species do not develop wings, despite having the wing genes, genome editing is very useful in investigating the involvement of enhancers and promoters.
One gene regulates the expression of another
We also know that wing genes and genes that are expressed in the legs are active in the formation of the long horn of the Japanese rhinoceros beetle (Trypoxylus dichotomus) (Figure 4). Genes do not function in isolation in the first place and if a gene is expressed in a different place or at a different time from what would usually be expected, they often curb the expression of other genes. When that happens, something will unexpectedly occur in a completely different location. We are learning that even when it seems as though a new phenomenon has occurred, it turns out actually to have been due to an existing network of genes.

Figure 4. Japanese rhinoceros beetleIt has been discovered that a wing formation gene and a leg formation gene are involved in the formation of a beetle’s horn. New insights show how the gene is involved in wing formation.
Genome editing is very useful for finding out the roles of such DNA regions that do not encode proteins. We are now able to investigate their role by cutting out or increasing those DNA regions themselves.
Let us take an example, although it does not relate to insects. Snakes have a gene called sonic hedgehog, which relates to leg formation. It has already been reported that snakes do not develop legs because part of the region that regulates this gene is missing. It has been reported that using genome editing to alter the region of sonic hedgehog involved in regulation to the pattern found in snakes actually inhibits leg formation. This offers direct proof that removing the region that causes a gene to be expressed affects the phenotype, even if the gene that encodes proteins is present. Previously, techniques like this could only be used in a limited number of model organisms, but genome editing has made it possible to apply them in virtually all organisms.
As mentioned above, even apterygote insects, which do not have wings, have the genes required to make wings in fruit flies. This suggests the possibility that it was the regions that regulate how those wing genes are expressed that were key to the acquisition of wings. Using genome editing techniques will enable us to directly research the role of those regulating regions and, for example, edit pterygote genes to fit the apterygote pattern, or vice versa. As we will also see progress in hitherto-impossible studies focused on gene networks, it will become possible to conduct research that takes us even closer to unraveling the mystery of wings.