Since peptide synthesis was first established in the 1950s, techniques have continuously evolved and broadened the application of peptides in a wide range of manufacturing settings, including the development of various materials and products, and in drug discovery. The diverse variety of items that can be manufactured because of peptide synthesis include gold (Au) nanoparticles synthesized using peptides for use in photothermal therapy, which involves heating cancer cells to a specific temperature to kill them, cell culture interfaces used in regenerative medicine, and sensors to detect viruses and explosives.
Special Feature 1 – The World of Peptides The multifarious items manufactured with the aid of peptide synthesis
composition by Rie Iizuka
illustration by Koji Kominato
I am sure that many readers think of peptides as nutrients composed of amino acids that are essential to the formation of muscles, internal organs, and other parts of the body. In addition to their nutritional properties, peptides are also being synthesized artificially and widely used in a variety of manufacturing settings.
One example is the synthesis of metals in a manner similar to biomineralization. Biomineralization refers to the process by which living organisms crystallize minerals to form structures, such as human teeth and bones. This process is also used by some species of microorganisms to create incredibly elaborate forms such as coccoliths in coccolithophores, silicic acid in diatoms, and micro-magnets in magnetic bacteria. Peptides are known to be involved in this mechanism.
In recent years, it was found that peptides can crystallize rare earths and other rare metals; many findings relating to biomineralization using peptides have been reported.
We also conducted a great deal of research to investigate the peptides that crystallize Au ions.
A number of peptides thought to be associated with Au nanoparticle (AuNP) binding have already been identified. Therefore, we first analyzed their amino acid composition, combined these amino acids to design randomly differing peptide sequences, and then compared their respective performances in binding with Au.
From this, we discovered that the key peptides were an amino acid sequence sandwiched between tryptophan (W), a sequence sandwiched between histidine (H) and W, and a sequence sandwiched between W and H (Figure 1).
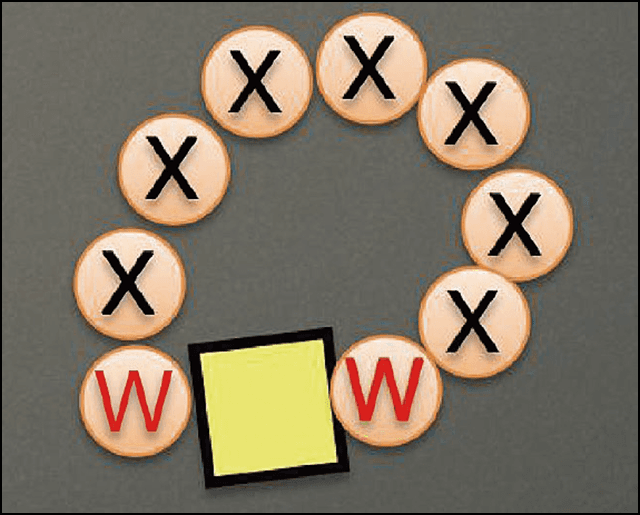
Figure 1. Peptide sequences used for generating AuNPsA sequence with tryptophan (W) as its base was identified in 46 of the 50 peptides with a high affinity for binding with AuNPs. X represents any amino acid. In many cases, histidine (H) was also present.
Could this be applied to photothermal cancer therapy?
Next, when we added Au ion solution to several peptides with a high binding affinity to Au, the solutions displayed a variety of colors, including yellow and red. This display of color is attributable to the differences in surface plasmon resonance corresponding to the shape of the AuNPs which was influenced by the type of peptide used. Remarkably, we found that the sequences of peptides used were involved in shaping the gold nanoparticles and that their sizes and shapes differed with different sequences (Figure 2).
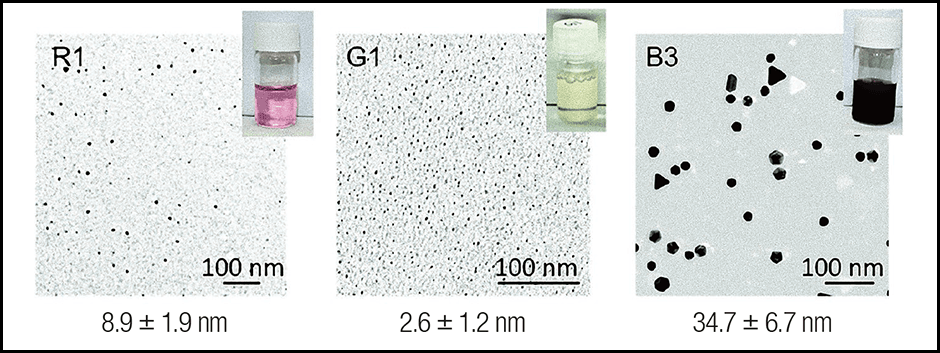
Figure 2. Particles differ according to the synthesized peptideColor differences in peptide bound AuNPs. It is thought that differences in the shape of the bonded AuNPs caused the color of the solution to differ according to which peptide had been synthesized. AuNPs in the red solution measured approximately 10 nm (1 nm is 1 billionth of a meter), while those in the yellow measured approximately 2–3 nm, and the bluish solution contained very large-diameter particles shaped like prisms and decahedrons.
The crucial amino acid with a major influence on the particle shape was W (Try) and H (His). While the red and yellow solutions contained a large amount of W and H, the peptides in the solution with larger gold nanoparticles tended to contain low amount of H. Of the nanoparticles created in this manner, we hypothesize that the near-infrared absorbing triangular Au nanoplates (TrAuNPls) produced using a peptide called B3 have potential applications in photothermal cancer therapy.
Taking advantage of the fact that cancer cells are more vulnerable to heat than normal cells, photothermal therapy kills cancer cells via the administration of a photothermal material that generates heat when exposed to light. Conventional techniques for treating cancer include surgery, chemotherapy, and radiotherapy. Photothermal therapy is a novel option that imposes less burden to the patient. We believe that we successfully synthesized AuNPs that can be used as photothermal therapy materials.
Although surfactants are usually used to control crystallization during the synthesis of Au nanoparticles, safety is crucial if they are to be used in living organisms such as in the case of photothermal therapy. Peptides are thought to be more biocompatible than surfactants, and tests have shown that using peptides to synthesize AuNPs results in a higher cellular survival rate than the use of surfactants.
After confirming that the AuNPs created using the B3 peptide heat up when irradiated with near-infrared light, we conducted tests in which the nanoparticles were mixed with cancer cells and then irradiated with near-infrared light. The results confirmed that the cells died (Figure 3). A further step toward the practical application of this therapy will be achieved once the peptides added can be equipped with a mechanism tailored specifically to cancer cells.
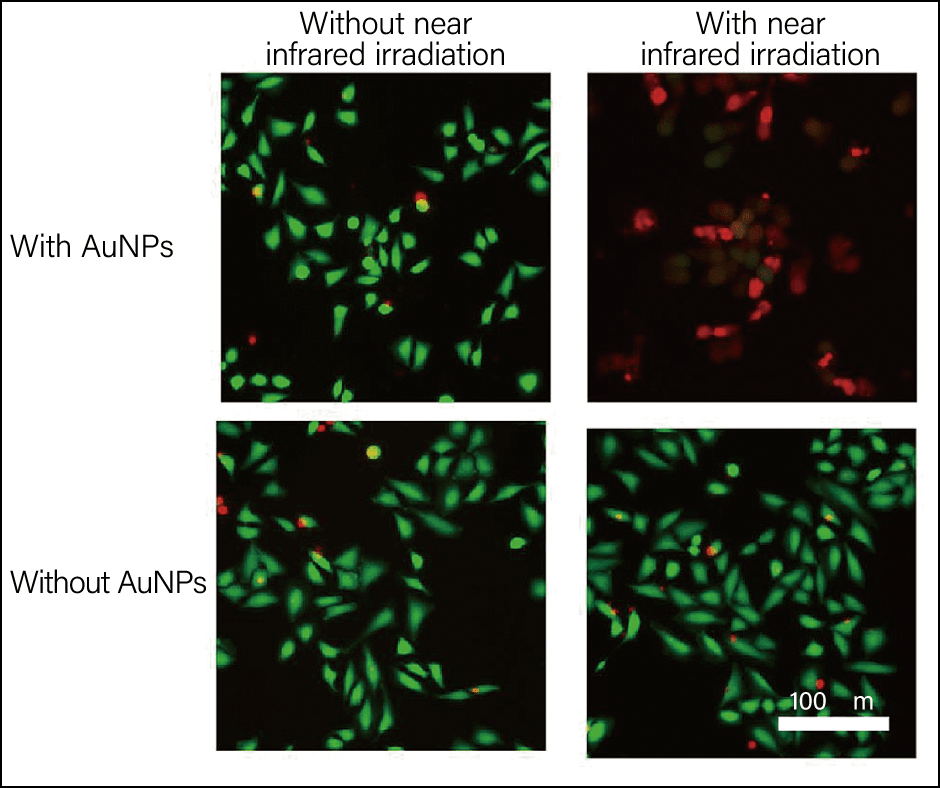
Figure 3. Cancer cells mixed with AuNPs and irradiated with near-infrared lightThe images on the left show cells not exposed to NIR (near infrared) irradiation and those on the right show cells that have been exposed. The top row of the images shows the situation where triangular nanoplates are present, while the bottom row shows the situation in their absence. Green indicates living cells, and red indicates dead cells.
Using peptides to make sensors
Allow me to introduce an additional example of the use of peptide synthesis in manufacturing. We participated in a program entitled “Ultra-high Speed Multiplexed Sensing System Beyond Evolution for the Detection of Extremely Small Quantities of Substances,” led by the program manager Reiko Miyata at Nagoya University, under the umbrella of the Impulsing Paradigm Change through Disruptive Technologies (ImPACT) Program. Professor Miyata’s program involved the use of peptides to make sensor components. We designed peptide probes (sensor components) capable of recognizing molecules that can be fitted to various devices.
Peptides have some degree of molecular recognition capability, and they are also more stable than proteins since denaturation does not occur with short peptides that do not form secondary structures. This makes peptides easier to manage from a quality control perspective. In addition, as their surfaces can be modified in a self-organizing manner and can also be chemically modified, peptides are simple to handle, so we believed that they could readily be applied in sensors used in everyday settings.
- *1 Self-organizing:Creating an orderly arrangement overall, even though there is no system controlling the overall arrangement.
As part of the aforementioned program, we designed peptide probes tailored to detect a variety of substances, including bacteria, viruses, PM2.5 particulate matter, explosives, smells, and stress markers (Figure 4).
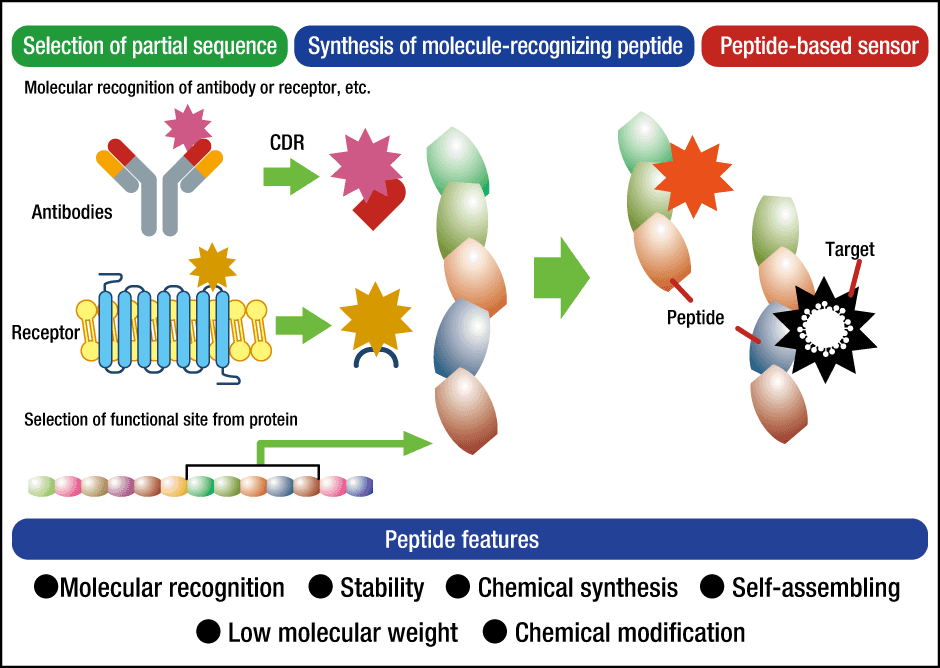
Figure 4. Peptide probe capable of molecular recognitionAntibody-based design of a peptide with the ability to recognize a target molecule, for the purpose of using the peptide in a sensor.
We began working on a peptide for detecting viruses.
Antibodies to viruses are proteins with a very powerful ability to recognize molecules. The part of an antibody’s structure, called the variable region, is known to be involved in antigen-specific recognition of substances. Within the variable region, areas at the tips called complementarity-determining regions (CDRs) react most sensitively to antigens, producing antibodies by rearranging amino acids. Assuming that peptides in these regions had the ability to recognize molecules, we began to investigate them.
First, we designed a probe to distinguish between type A and B influenza. Having designed a peptide with a high affinity for binding with hemagglutinin (HA), which is used to distinguish type A influenza from type B, we are then developing a sensor that would identify whether an influenza virus is type A or type B. At present, simple tests used to detect the influenza virus cannot distinguish the types unless the virus has sufficiently multiplied within the body. We know that this sensor offers a very high rate of identification even when only 10 or so viral particles pass through it. As such, we hope it will enable drug administration to begin at an earlier stage than is currently possible.
Peptides capable of binding to bacteria
To search for peptides capable of binding with bacteria, we examined toll-like receptors (TLRs), which recognize microbial patterns. We used the amino acid sequences of these TLRs to search for peptide portions that seemed likely to be responsible for recognizing the relevant antigens. These TLRs include TLR2, which recognizes peptidoglycan, which is often seen in the cell wall of gram-positive bacteria; TLR5, which recognizes flagellin, a structural protein found in microbial flagella; and TLR4, which recognizes lipopolysaccharide (LPS), a component present in the cell wall of gram-negative bacteria.
When we synthesized a peptide based on TLR4 and investigated its affinity for binding with the bacterium Escherichia coli (E. coli) and LPS, our findings suggested that the sequence observed in the bond demonstrated a very high recognition of both targets.
As the ultimate purpose of searching for this peptide is to develop a probe for a device, it is vital that it functions when combined with the main sensor unit.
We conducted an experiment in which we introduced microorganisms into a cavity on the surface of a silicon nitride film to observe the difference in electrical current (Figure 5). The pore sensor (sensor device) was kindly provided by Associate Professor Makusu Tsutsui, Professor Masateru Taniguchi, Specially Appointed Professor Tomoji Kawai, and Professor Takashi Washio of SANKEN (formerly known as the Institute of Scientific and Industrial Research), Osaka University, in a joint research project with our team. When we installed a synthetic peptide capable of recognizing flagellin, we discovered a protein found in the flagella of microorganisms inside the hole. When both a wild strain of E. coli and E. coli lacking flagellin were encountered, the sensor reacted strongly to the wild strain of the bacterium. This shows that it is possible to detect a target substance by designing a peptide tailored to the features of the target.
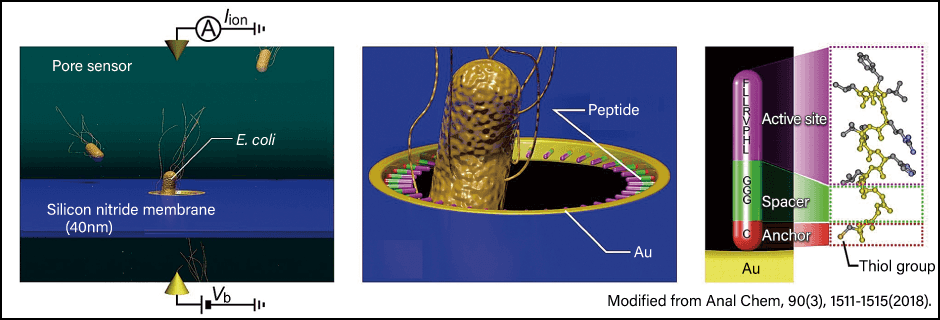
Figure 5. Sensor mechanismThe bacterium or other target passes through a hole in the pore sensor. The peptide probe installed in the hole consists of an inner peptide that detects the target, a peptide bound to the material used on the hole side, and a peptide in the middle that connects the two.
We are also developing a sensor for cancer exosomes (extracellular vesicles) as part of a joint research project with Associate Professor Takao Yasui and Professor Yoshinobu Baba of Nagoya University.
The exosomes released from cancer cells promptly enter blood vessels and assist in metastasis of the cancer cells due to their signaling factors. Accordingly, they have recently begun to attract attention due to their potential in cancer treatment, diagnosis, metastasis prediction, and postoperative prognosis prediction. We believe that it might be possible to produce an exosome-based diagnostic device using a sensor fitted with a peptide that recognizes these exosomes.
Tetraspanin CD9 is known to be an exosome marker protein, so we designed a peptide to recognize it and this peptide was used in device construction. Our experiments showed that the exosomes were properly detected.
Antibody-based tests are also possible, but the interaction of antibodies with exosomes is substantial, making it difficult to elute the captured exosomes. An advantage of peptides is that we can harvest exosome concentrates for cases in which other biogenic substances are reduced.
We believe that it might be possible to deploy this CD9-binding peptide not only for diagnosis but also for future treatments. This is because the CD9-binding peptide that we synthesized might be able to obstruct or alter the functions of CD9. Our tests also found that when this CD9-binding peptide was added to a plate on which densely packed cancer cells were grown, the migration of the cancer cells in that area was suppressed in a concentration-dependent manner. In other words, we might be able to suppress cell migration and infiltration capabilities in cells with high expression of the exosome marker CD9.
Various micromachined devices
Peptides can be used in more applications than just as a sensor probe for living organisms. Kyushu University Research Professor Kiyoshi Toko has created an antibody capable of recognizing explosives and used it to develop a sensing system. We investigated the area of the antibody that recognizes trinitrotoluene (TNT). We found that the peptide we discovered could also detect TNT.
As DNA aptamers bind not only with enzymes, receptors, and viral proteins, but also with metal ions, research has focused on creating a recognition device for a sensor that applies the mechanism via which DNA recognizes various molecules. We believe that peptides can also be used in a similar manner. In fact, as the amino acids that could be used are more numerous than DNA, we hope that it might be possible to achieve a wide range of recognition capabilities.
- *2 Aptamer: A nucleic acid that binds specifically to a certain substance. Aptamers inhibit cell and protein function.
A variety of micromachined devices have been created using nanotechnology, but they face challenges in ensuring the biocompatibility of their surfaces. In the field of biosensing, the question of how to build an interface is crucial. As peptides can be used as sensing molecules to detect various substances and are easy to handle, it is hoped that they might function as interfacial molecules in the future.