Bacteriophages are viruses that infect bacteria. As almost all infected bacteria die, phages could be described as the natural predators of bacteria. The treatment of infectious diseases using this effect is called phage therapy and was once very common. However, due to the discovery of antibiotics, phage therapy eventually became a forgotten approach. In recent years, though, phage therapy has come back under the spotlight, with the emergence of multidrug-resistant bacteria, against which antibiotics are completely ineffective.
Special Feature 1 – The Unknown World of Viruses Bacteriophages: the natural predators garnering attention with the emergence of multidrug-resistant bacteria
composition by Rie Iizuka
Bacteriophages (referred to below as “phages”) are viruses that infect bacteria. Their existence was known of as early as 1915 (Figure 1). As the infected bacterium appears to be swallowed up when its cell membrane is destroyed and it is killed (a process known as bacteriolysis), these viruses were given the name bacteriophage —— meaning “that which eats bacteria” —— derived from the Greek word phagos (eater).
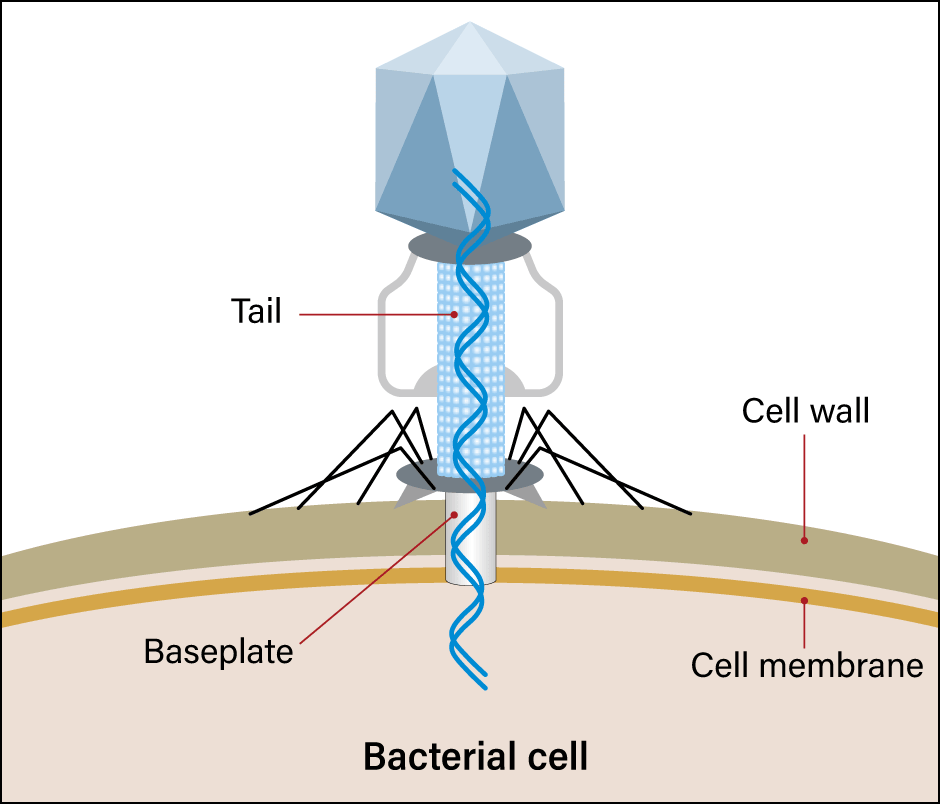
Figure 1. Phage infectionMany phages are found in the environment. They cling to bacteria and inject nucleic acid into them. The nucleic acid is replicated in the bacterium, eating away at the bacterium until it ultimately emerges.
Multidrug-resistant bacteria cause almost as many deaths as cancer
Phages were discovered in the early 20th century, when irrigation and flood control systems had not yet been developed and death rates from bacterial diarrhea and the like were high. While phages were used to treat such diseases, they became completely obsolete in Western countries due to the discovery of penicillin in 1928, after which antibiotics reigned supreme in the West. However, for some time during the Cold War, the Eastern Bloc was unable to obtain the new antibiotics developed in the West, so they developed phages to take the place of antibiotics in various industries.
For example, phages that kill the microbes that cause fruit and vegetables to rot are sprinkled over produce like a food additive. In addition, they are used on meat and fish sold by the piece at markets, and drugs featuring a cocktail of different phages can also be bought, marketed in a similar way to Lactobacillus preparations and other such intestinal regulators in Japan. Other products include phage-based acne treatments and disinfectants for floors at medical facilities (Figure 2).
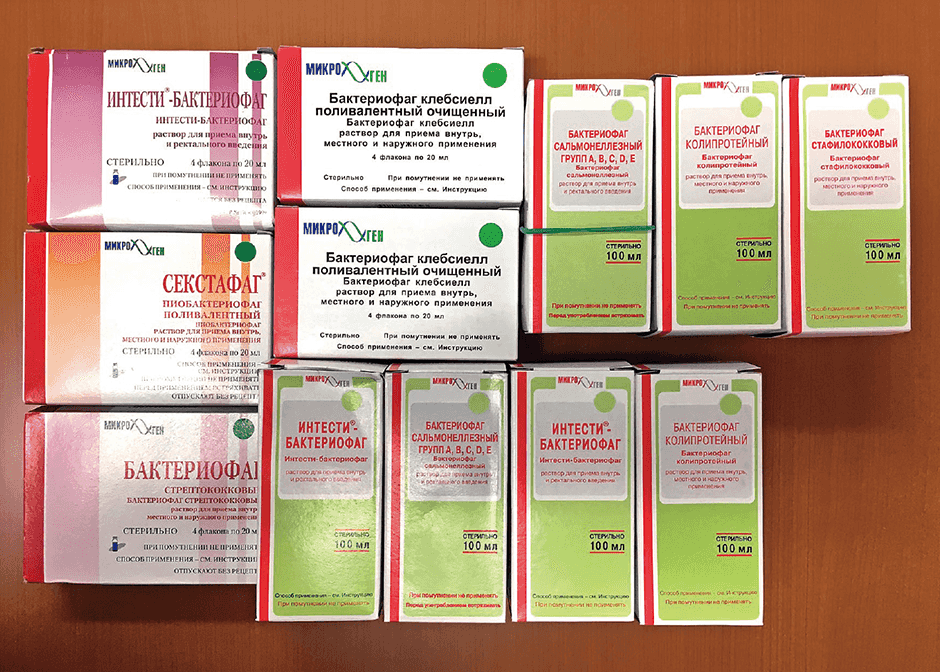
Figure 2. Phage drugs on sale in RussiaRanging from medications requiring a physician’s prescription to those readily purchased directly from a local pharmacy, phage drugs are widely used in Russia.
Since the end of the Cold War, multidrug-resistant bacteria have gradually begun to appear in the East, as well, due to the worldwide abuse of antibiotics. Multidrug-resistant bacteria are no longer a problem confined to the West. The World Health Organization (WHO) estimates that, across the globe, multidrug-resistant bacteria could cause 10 million deaths each year by 2050, overtaking cancer as a cause of death.
The Patterson case triggered worldwide interest in phages. In 2016, Tom Patterson, a professor at the University of California, San Diego, developed a pancreatic infection caused by a multidrug-resistant strain of Acinetobacter baumannii during a trip to Egypt. Existing antibiotics proved completely ineffective, and Patterson hovered between life and death.
Patterson’s wife, who was also a scientist at the university, investigated phage therapy and the medical team decided to try it. However, there is no one-to-one correspondence between phages and the bacteria they kill —— that is to say, it is not the case that phage A will basically kill bacterium B. The treatment process began by screening phages that cause bacteriolysis in the bacterium with which Patterson was infected. His physicians eventually found effective phages among the stocks of the U.S. military and research facilities overseas. They proved remarkably efficacious and brought Patterson back from the verge of death.
Research institutes worldwide are focusing on phages
Research institutes across the globe began to focus their attention on phages as a result of this epoch-making case. Soon after the Patterson case, a patient at a London hospital who was severely immunocompromised due to leukemia developed a multidrug-resistant atypical mycobacterial infection and was treated with genetically modified phages. There are hopes for new treatments using phages to address conditions other than bacterial infections as well, with phages having been shown to be capable of treating alcoholic liver disease in mice by causing the bacteriolysis of specific intestinal bacteria.
Research into phage therapy is also beginning to become more widespread in Japan. In undertaking research into gut immunity, I aimed to use phages to treat diseases caused by specific intestinal bacteria.
First, it was necessary to start by searching for intestinal bacteria phages. Phages that coexist with us while infecting bacteria are always present in our gut. However, when I first embarked on this research, culturing anaerobic intestinal bacteria was in itself difficult, while isolating the phages that infected them was a nigh-on impossible task.
Even in 2015 or thereabouts, any phage-like sequences identified by next-generation sequencers were listed as “unknown” in the database, to the extent that intestinal phages were referred to as “viral dark matter.” We succeeded in establishing a technique for collecting phage genomes and DNA from feces, in order to advance the search for intestinal phages. After using next-generation sequence analysis to collect a comprehensive range of phage genome fragments, we reconstructed the original phage base sequences with the aid of a supercomputer. We then managed to create a pipeline (a term used in fields such as computing to refer to a set of data processing elements connected in series, where the output of one element is the input of the next one) that enabled us to classify them by determining their gene regions and cloning the viruses on the computer.
In 2020, we used this to publish a database of the intestinal bacteria and phages of 101 healthy Japanese individuals (Figure 3). This is the first time in the world that the gut microbiota and intestinal phageome have been simultaneously classified from the same fecal samples. As a result, we discovered that the intestinal phageome can be divided into as many as three patterns.
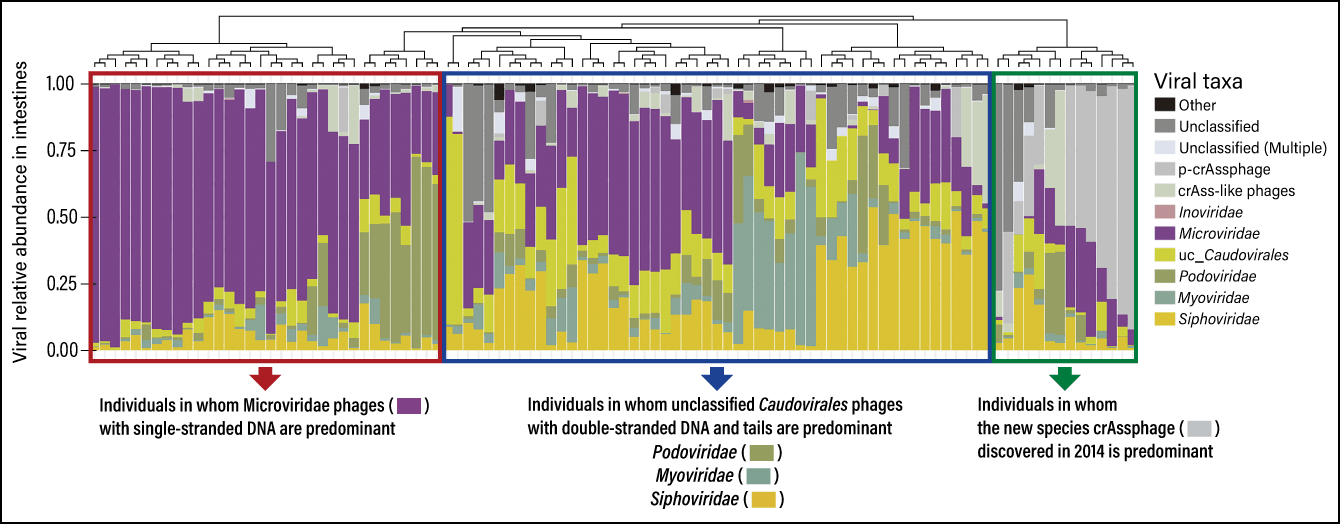
Figure 3. Intestinal phageome composition of healthy Japanese individualsThe individuals were divided into three groups according to the predominance of phages in their intestines: those with mostly Microviridae, which have a single-stranded DNA genome; those with mostly Caudovirales (Podoviridae, Myoviridae, Siphoviridae, etc.), which have a tail; and those with mostly crAssphages. CrAssphages were isolated in 2014 as a new species of phage. Many phage families with similar structures and functions to crAssphages have been found not only in humans, but also in livestock, rodents, and the environment.
Although there may be differences due to individual physical condition and the like, we had the impression that the predominant phage groups are fixed, as the intestinal phageome has less variation in species than the intestinal microbiota. We have not yet been able to unravel any correlations between phage patterns and diseases. However, say that Phages α, β, and several others are capable of infecting Bacterium Z, and Subject A’s gut contains a large number of Phage α, this would mean that Phages α and β vied with each other for a host, in the form of Bacterium Z, and Phage α has prevailed in the intestinal environment of Subject A. This is thought likely to be something inherited from the individual’s mother or determined by environmental factors. We want to undertake further research into this area in due course.
We can see which intestinal bacteria are infected
At the same time, we developed two techniques for illustrating host bacteria-phage associations. One applies the bacterial immune system known as the CRISPR-Cas system, which is the focus of attention in the field of genome editing.
When bacteria are first infected by a phage, an enzyme called Cas bites off an important sequence from the phage that is distinctive and hard to alter, and incorporates it into its own CRISPR gene locus (spacer). In other words, these sections could be described as a record of the bacteria’s past infections. If that is the case, comparing this section with phages in a database might show us that the phage sequence incorporated came from a siphovirus, for example.
And if we search for the types of bacteria with a spacer corresponding to siphovirus, we will be able to see which intestinal bacteria are infected with siphovirus. So far we have learned that Bacteroidetes —— one of the most prevalent bacterial phyla in the gut microbiota —— has a distinctive spacer sequence derived from crAssphage, which was discovered within the last decade. This means that, at the very least, crAssphage has infected Bacteroidetes.
The other search method stems from phage infection patterns. There are two types of phages: lytic, which completely kill bacteria when they infect them, and lysogenic, which coexist with host bacteria after being integrated into them when a genome is inserted into the bacterium’s circular chromosome (Figure 4). The integrated portion is called a prophage. We can find out which bacteria integrated the prophage by conducting homologous analysis of these portions with a database of intestinal bacteria genomes. We developed the ability to search for lysogenic phages in a single sweep, most notably searching for sequences that were a 100% match for Bacteroides uniformis. Thus, we were able to compile a database of which phages infect which bacteria.
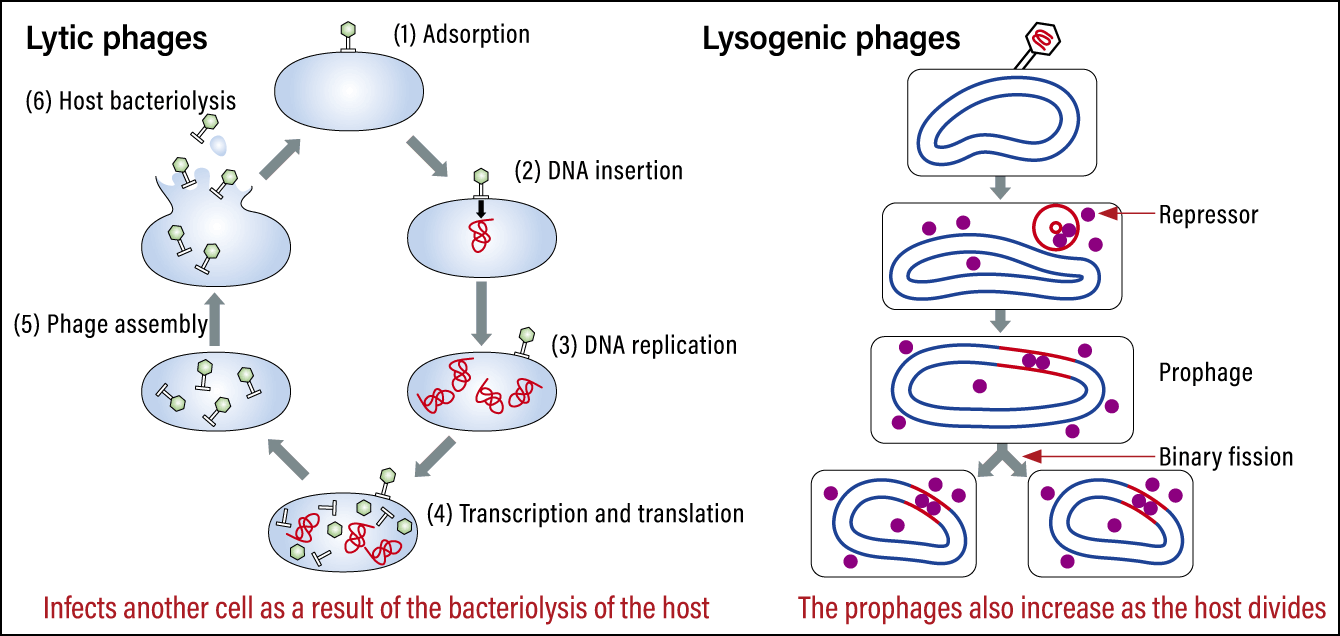
Figure 4. Lytic phages and lysogenic phagesLytic phages destroy the cell and then infect the next cell. Lysogenic phages are integrated into the cell they effect and increase as the cell divides.
We are now undertaking research aimed at leveraging these results in therapies, focusing first of all on treating pseudomembranous colitis.
Pseudomembranous colitis is a disease that occurs when the balance of intestinal bacteria is disrupted by the administration of antibiotics, enabling Clostridioides difficile —— normally present only in small amounts —— to proliferate and produce toxins that cause colitis. In most cases in Japan, the normal flora in the gut regenerate and exterminate the C. difficile once treatment with the initial antibiotics is stopped, but highly virulent strains with a binary toxin described as a third toxin have been discovered overseas. These strains are multidrug-resistant, causing death from severe pseudomembranous colitis in many cases.
However, one feature of C. difficile is its resistance to phage infection and no lytic phages have been discovered. Accordingly, we focused on phage-derived enzymes.
Phage-derived enzymes under the spotlight
One of the best-known types of phage-derived enzymes are proteins called endolysins. Produced by phages in bacteria, endolysins cleave and destroy the peptidoglycan layer of the bacterial cell membrane. However, in the case of gram-positive bacteria, it is also possible to destroy the cell membrane by sprinkling this enzyme on the bacteria from the outside, because the peptidoglycan layer is exposed. Due to the possibility that they might be suited to such uses as treating mastitis in dairy cows, in which it is preferable to refrain from using antibiotics, phage-derived enzymes have attracted attention in a variety of fields. We thought of putting these enzymes to use because, as luck would have it, C. difficile is a gram-positive bacterium.
We analyzed the genomes of C. difficile found in the gut microbiota of healthy individuals and the genomic data of C. difficile isolated from patients who had developed pseudomembranous colitis. As a result, we succeeded in finding a number of lysogenic phages that had been integrated into C. difficile as prophages. When we then searched for the genome of endolysin with bacteriolytic capacity against C. difficile, we found 10 different types. When we synthesized the proteins in accordance with these sequences and tested them, we found that a number of enzymes performed very well in suppressing C. difficile.
Lysogenic phages that produce prophages are known to travel between bacteria. For example, the Verotoxin produced by Escherichia coli O157 is genetically very similar to the Shiga toxin produced by Shigella dysenteriae, so it is believed to have been propagated by the same lysogenic phage. Accordingly, while the therapeutic use of phages derived from lysogenic phages had been deemed to be contraindicated, we believe that focusing on enzymes rather than phages and synthesizing the enzyme portion alone could open up possibilities for new treatments.
Naturally, it is envisaged that some bacteria will become resistant to phages, as they have to antibiotics. To counteract this issue, it is recommended that phage therapy should employ a cocktail of phages, rather than just a single one. It is also hoped that their applications could go beyond bacteria associated with infectious diseases, with the host specificity of phages being used to suppress specific intestinal bacteria with pinpoint accuracy, without affecting useful bacteria.
Furthermore, we are exploring a variety of other therapies, including genetically modified phages and next-generation phage therapy based on synthesizing the lytic enzymes produced by phages and using them as antibiotics.
There are high barriers to the commercialization of some items in Japan, but if exchanges with other countries flourish again once the dust settles in the wake of the COVID-19 pandemic, one cannot deny the possibility that Japan might face a mass influx of multidrug-resistant bacteria that are not currently a major problem here. To prepare for such an eventuality, I want to equip us with as many phage therapy options as possible. I would like to bring to fruition the advanced phage medicine of the future, using phages to conquer multidrug-resistant bacteria and create new therapies by suppressing intestinal bacteria, among others.